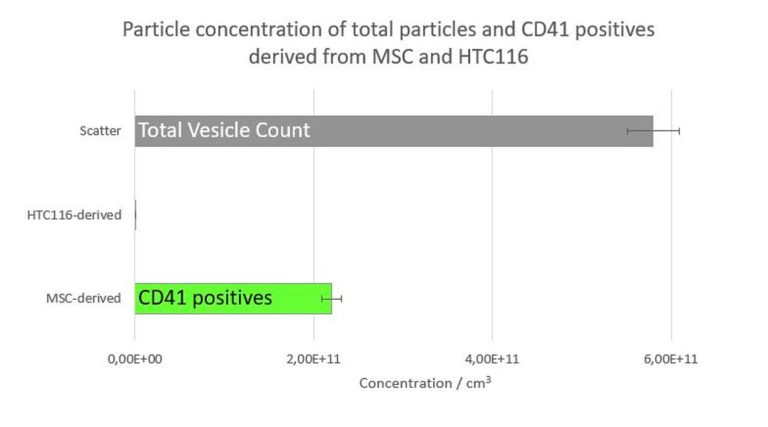
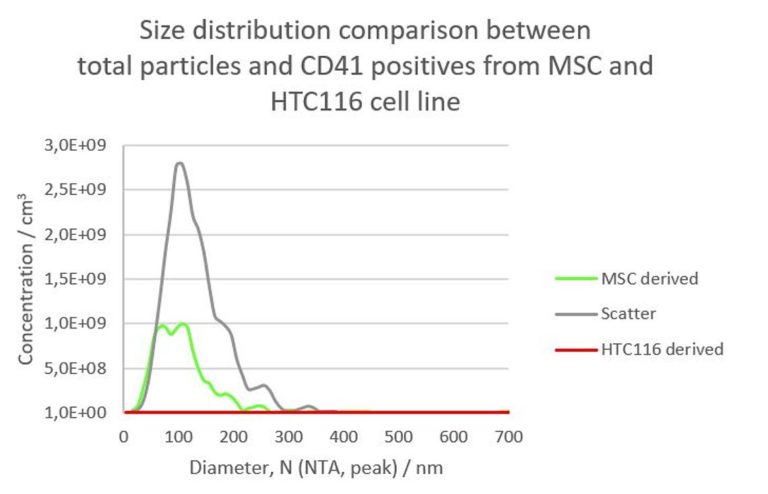
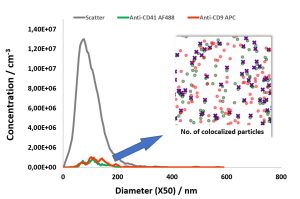
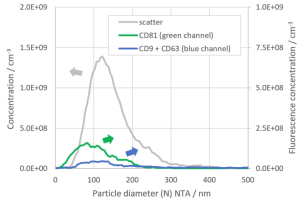
Baixauli F., Lopez-Otin C., Mittelbrunn M. Exosomes and autophagy: coordinated mechanisms for the maintenance of cellular fitness. Front Immunol. 2014; 5:403.
Becker A., Thakur B., Weiss J. M., Kim H. S., Peinado H., Lyden D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell. 2016 Dec 12;30(6):836-848.
Console L., Scalise M., Indiveri C. Exosomes in inflammation and role as biomarkers. Clin Chim Acta. 2019; 488:165-171.
Deatherage B.L. and Cookson B.T. Membrane Vesicle Release in Bacteria, Eukaryotes, and Archaea: a Conserved yet Underappreciated Aspect of Microbial Life. Infect Immun. 2012; 80(6):1948-57.
Giebel B, Helmbrecht C. Methods to Analyze EVs; Methods Mol Biol. 2017;1545:1-20.
Konoshenko M.Y., Lekchnov E.A., Vlassov A.V., Laktionov P.P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. Biomed Res Int. 2018: 8545347.
Johnstone R. M., Adam M., Hammond J. R., Orr L., Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). The Journal of Biological Chemistry. 1987; 262 (19): 9412–20
Mathivanan S., Ji H., Simpson R.J. Exosomes: extracellular organelles important in intercellular communication. J Proteom. 2010;73(10):1907–1920.
Rahbari M., Pecqueux m:, Aust D., Stephan H., Tiebel O., Chatzigeorgiou A., Tonn T., Baenke B., Rao V., Ziegler N., Greif H., Lin K., Weitz J., Rahbari N. N. and Kahlert C. Expression of Glypican 3 Is an IndependentPrognostic Biomarker in Primary Gastro-Esophageal Adenocarcinoma and Corresponding Serum Exosomes. J Clin Med. 2019 May 16;8(5).
Raposo G., Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383.
Record M., Carayon K., Poirot M., Silvente-Poirot S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim Biophys Acta. 2014;1841(1):108–120.
Rupert, D. L. M., Claudio, V. Lässer, C. Bally, M. Methods for the physical characterization and quantification of extracellular vesicles in biological samples. Biochim Biophys Acta Gen Subj. 2017; 1861(1 Pt A):3164-3179.
Soo C.Y., Song Y., Zheng Y., Campbell E.C., Riches A.C., Gunn-Moore F. and Powis S.J. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology. 2012, 136:192–197.
Stanly C., Fiume I., Capasso G., Pocsfalvi G. Isolation of Exosome-Like Vesicles from Plants by Ultracentrifugation on Sucrose/Deuterium Oxide (D2O) Density Cushions. Methods Mol Biol. 2016; 1459:259-69.
Weber A., Wehmeyer J. C., Schmidt V., Lichtenberg A., Akhyari P. Rapid Fluorescence-based Characterization of Single ExtracellularVesicles in Human Blood with Nanoparticle-tracking Analysis. J. Vis. Exp. 2019 (143), e58731
Yanez-Mo M., Siljander P.R., Andreu Z., Zavec A.B., Borras F.E., Buzas E.I., Buzas K., Casal E., Cappello F., Carvalho J., Colas E., Cordeiro-da Silva A., Fais S., Falcon-Perez J.M., Ghobrial I.M., Giebel B., Gimona M., Graner M., Gursel I., Gursel M., Heegaard N.H., Hendrix A., Kierulf P., Kokubun K., Kosanovic M., Kralj-Iglic V., Kramer-Albers E.M., Laitinen S., Lasser C., Lener T., Ligeti E., Line A., Lipps G., Llorente A., Lotvall J., Mancek-Keber M., Marcilla A., Mittelbrunn M., Nazarenko I., Nolte-’t Hoen E.N., Nyman T.A., O’Driscoll L., Olivan M., Oliveira C., Pallinger E., Del Portillo H.A., Reventos J., Rigau M., Rohde E., Sammar M., Sanchez-Madrid F., Santarem N., Schallmoser K., Ostenfeld M.S., Stoorvogel W., Stukelj R., Van der Grein S.G., Vasconcelos M.H., Wauben M.H., De Wever O. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015; 4:27066.